
Transportability Analysis with cfperformance
Christopher Boyer
2026-01-28
Source:vignettes/transportability.Rmd
transportability.RmdOverview
This vignette demonstrates how to use the
transportability functions in
cfperformance to evaluate prediction model performance when
transporting from a source population (such as a randomized controlled
trial) to a target population.
The methods are based on Voter et al. (2025), “Transportability of machine learning-based counterfactual prediction models with application to CASS,” Diagnostic and Prognostic Research, 9(4). doi:10.1186/s41512-025-00201-y
When to Use Transportability Analysis
Transportability analysis is appropriate when:
- You have a prediction model trained in one population (e.g., an RCT)
- You want to evaluate performance in a different target population
- Treatment is randomized in the source population
- Covariates are measured in both populations
Common scenarios include:
- Evaluating an RCT-derived risk score in a real-world patient population
- Assessing whether a clinical prediction rule “transports” to a new setting
- Understanding how model performance varies across populations
The Setting
Consider two populations:
-
Source (S=1): Often an RCT where treatment
Ais randomized - Target (S=0): The population where we want to deploy the model
We observe: - Covariates X in both populations -
Treatment A in both populations
- Outcomes Y in the source (and possibly target) - Model
predictions g(X) for all individuals
The goal is to estimate the counterfactual prediction performance
— how well would the model perform in the target population if everyone
received treatment level a?
Using the Included Example Data
The package includes a simulated transportability dataset:
data(transport_sim)
head(transport_sim)
#> age biomarker smoking source treatment event risk_score
#> 1 0.63916439 -0.06780891 1 1 0 0 0.3388230
#> 2 0.06014812 0.81999552 0 1 0 0 0.3309058
#> 3 0.78997359 -1.00222911 1 0 1 0 0.2773807
#> 4 1.28410328 0.78155960 1 0 1 0 0.4481046
#> 5 0.39198673 1.24382106 0 1 1 0 0.3841622
#> 6 -0.13627856 0.61176294 1 1 1 0 0.3482827
# Population sizes
cat("Source (RCT) n =", sum(transport_sim$source == 1), "\n")
#> Source (RCT) n = 1276
cat("Target n =", sum(transport_sim$source == 0), "\n")
#> Target n = 1224The transport_sim dataset contains: - age,
biomarker, smoking: Patient covariates -
source: Population indicator (1 = source/RCT, 0 = target) -
treatment: Binary treatment (randomized in source,
confounded in target) - event: Binary outcome -
risk_score: Predictions from a model trained in the source
population
Transport Analysis
The transport analysis uses outcomes from the source/RCT to estimate performance in the target population. This is useful when:
- Outcomes are only observed in the RCT
- You want to leverage randomization in the source
MSE in the Target Population
mse_result <- tr_mse(
predictions = transport_sim$risk_score,
outcomes = transport_sim$event,
treatment = transport_sim$treatment,
source = transport_sim$source,
covariates = transport_sim[, c("age", "biomarker", "smoking")],
treatment_level = 0, # Evaluate under no treatment
analysis = "transport",
estimator = "dr",
se_method = "none"
)
print(mse_result)
#>
#> Counterfactual Transportable MSE Estimation
#> ---------------------------------------------
#> Analysis: transport
#> Estimator: dr
#> Treatment level: 0
#> N target: 1224 | N source: 1276
#>
#> Estimate: 0.2219
#>
#> Naive estimate: 0.2178Comparing Estimators
Let’s compare all four estimators:
estimators <- c("naive", "om", "ipw", "dr")
mse_estimates <- sapply(estimators, function(est) {
result <- tr_mse(
predictions = transport_sim$risk_score,
outcomes = transport_sim$event,
treatment = transport_sim$treatment,
source = transport_sim$source,
covariates = transport_sim[, c("age", "biomarker", "smoking")],
treatment_level = 0,
analysis = "transport",
estimator = est,
se_method = "none"
)
result$estimate
})
data.frame(
Estimator = estimators,
MSE = round(mse_estimates, 4)
)
#> Estimator MSE
#> naive naive 0.2178
#> om om 0.2222
#> ipw ipw 0.2213
#> dr dr 0.2219- naive: Uses source data with A=0 only (ignores population differences)
- om (outcome model): Models E[L|X,A,S=1] and averages over target X
- ipw: Reweights source observations to match target population
- dr (doubly robust): Combines outcome modeling and IPW
AUC in the Target Population
For discrimination, we can estimate the transportable AUC:
auc_result <- tr_auc(
predictions = transport_sim$risk_score,
outcomes = transport_sim$event,
treatment = transport_sim$treatment,
source = transport_sim$source,
covariates = transport_sim[, c("age", "biomarker", "smoking")],
treatment_level = 0,
analysis = "transport",
estimator = "dr",
se_method = "none"
)
print(auc_result)
#>
#> Counterfactual Transportable AUC Estimation
#> ---------------------------------------------
#> Analysis: transport
#> Estimator: dr
#> Treatment level: 0
#> N target: 1224 | N source: 1276
#>
#> Estimate: 0.6204
#>
#> Naive estimate: 0.6249ROC Curve in the Target Population
We can visualize the full ROC curve for the target population:
roc_result <- tr_roc(
predictions = transport_sim$risk_score,
outcomes = transport_sim$event,
treatment = transport_sim$treatment,
source = transport_sim$source,
covariates = transport_sim[, c("age", "biomarker", "smoking")],
treatment_level = 0,
analysis = "transport",
estimator = "dr",
include_naive = TRUE
)
# Plot the ROC curve
plot(roc_result)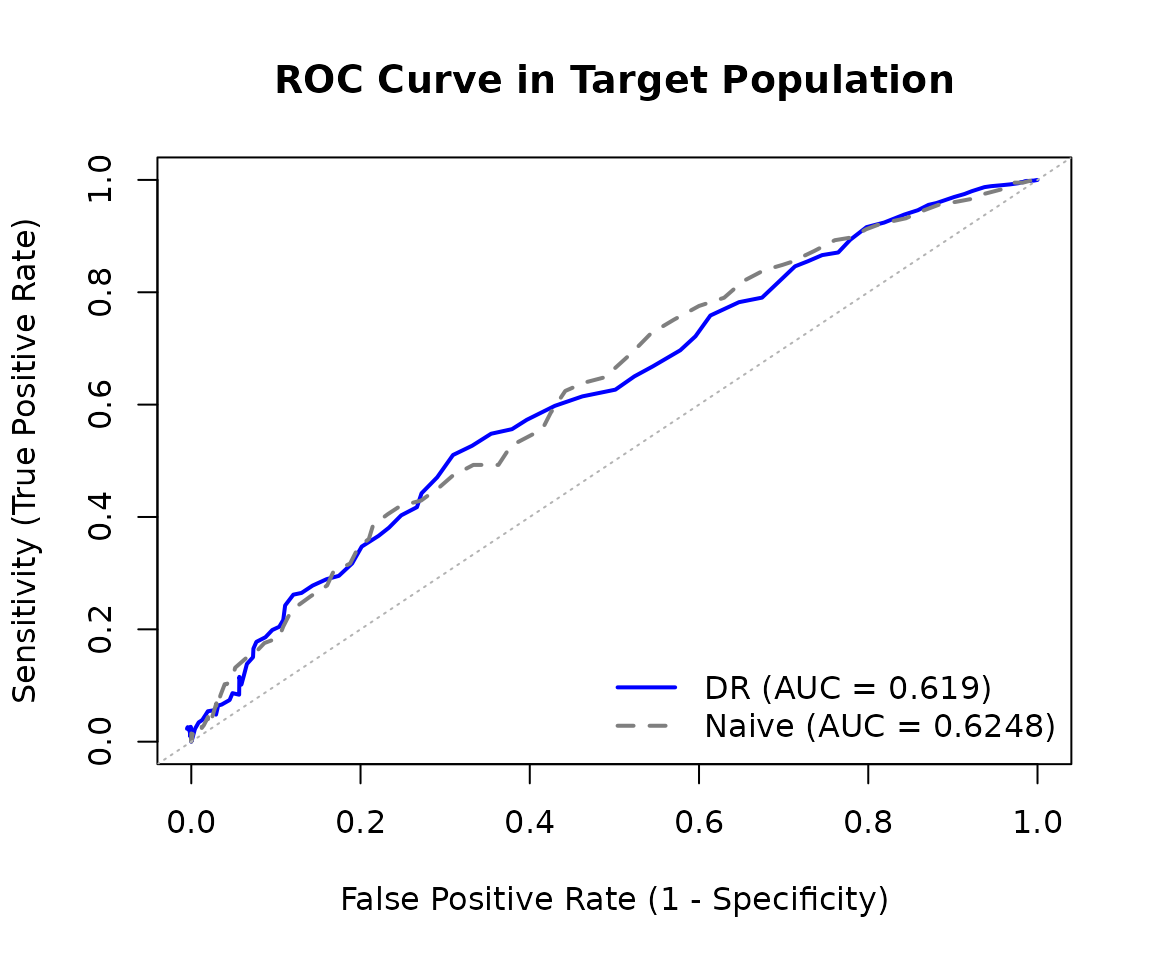
The comparison between the doubly robust and naive ROC curves shows how performance estimates change when properly accounting for the distribution shift between source and target populations.
Calibration in the Target Population
To assess calibration in the target population:
calib_result <- tr_calibration(
predictions = transport_sim$risk_score,
outcomes = transport_sim$event,
treatment = transport_sim$treatment,
source = transport_sim$source,
covariates = transport_sim[, c("age", "biomarker", "smoking")],
treatment_level = 0,
analysis = "transport",
estimator = "ipw",
smoother = "loess",
se_method = "none"
)
print(calib_result)
#>
#> Counterfactual Transportable CALIBRATION Estimation
#> ---------------------------------------------
#> Analysis: transport
#> Estimator: ipw
#> Treatment level: 0
#> N target: 1224 | N source: 1276
#>
#> Calibration Metrics:
#> ICI (Integrated Calibration Index): 0.0546
#> E50 (Median absolute error): 0.0558
#> E90 (90th percentile error): 0.0852
#> Emax (Maximum error): 0.1806
plot(calib_result)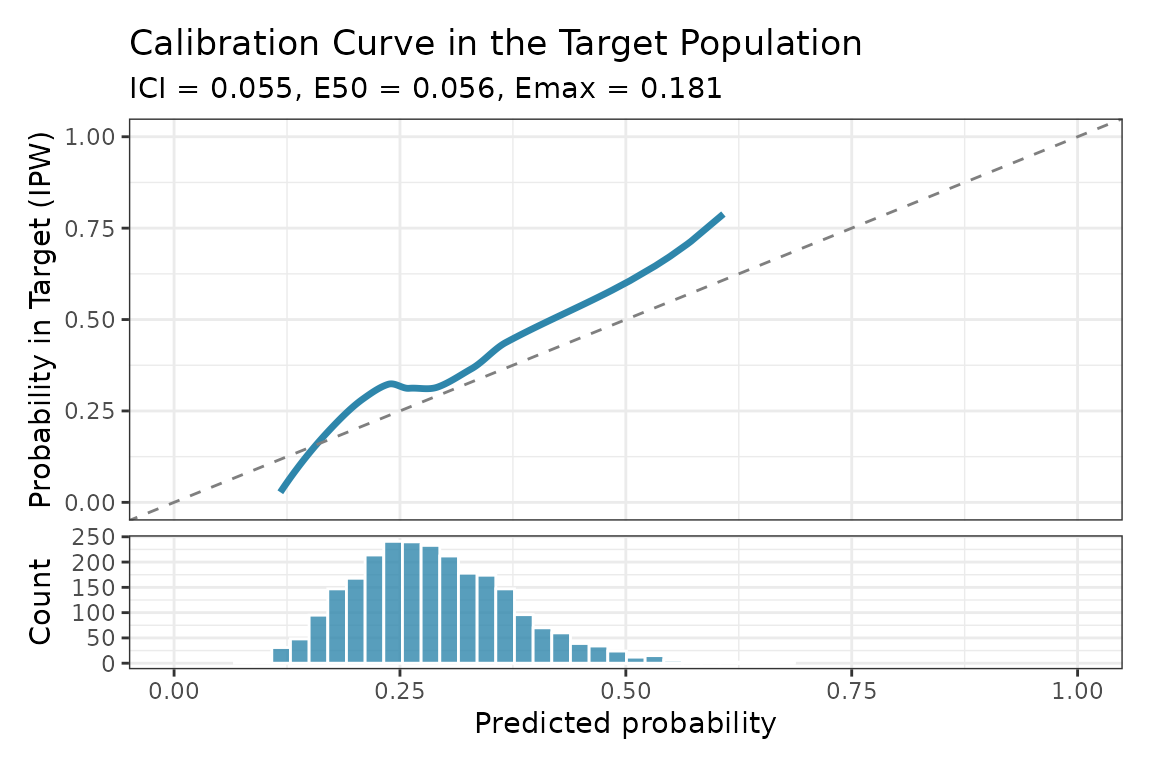
Transportable calibration curve for the target population
Joint Analysis
The joint analysis pools source and target data for potentially more efficient estimation. This requires outcome data in both populations.
mse_joint <- tr_mse(
predictions = transport_sim$risk_score,
outcomes = transport_sim$event,
treatment = transport_sim$treatment,
source = transport_sim$source,
covariates = transport_sim[, c("age", "biomarker", "smoking")],
treatment_level = 0,
analysis = "joint", # Pool data
estimator = "dr",
se_method = "none"
)
cat("Transport MSE:", round(mse_result$estimate, 4), "\n")
#> Transport MSE: 0.2219
cat("Joint MSE:", round(mse_joint$estimate, 4), "\n")
#> Joint MSE: 0.2223Factual Prediction Model Transportability
The examples above use counterfactual transportability, which estimates performance under a hypothetical treatment intervention. However, many prediction models are developed without a causal/counterfactual interpretation.
For factual prediction model transportability —
estimating how well a model predicts observed outcomes in the target
population — you can omit the treatment and
treatment_level arguments:
# Factual transportability (no treatment/intervention)
mse_factual <- tr_mse(
predictions = transport_sim$risk_score,
outcomes = transport_sim$event,
source = transport_sim$source,
covariates = transport_sim[, c("age", "biomarker", "smoking")],
# treatment = NULL (default),
# treatment_level = NULL (default),
analysis = "transport",
estimator = "dr",
se_method = "none"
)
print(mse_factual)
#>
#> Factual Transportable MSE Estimation
#> ---------------------------------------------
#> Analysis: transport
#> Estimator: dr
#> N target: 1224 | N source: 1276
#>
#> Estimate: 0.1892
#>
#> Naive estimate: 0.1948Notice the output shows “Factual Transportable” to indicate this mode.
Factual vs Counterfactual Mode
Factual mode (treatment = NULL):
- Estimates E[L(Y, g(X)) | S=0] — performance on observed outcomes
- Uses only the selection model P(S=0|X) for inverse-odds weighting
- No propensity score model needed
- Appropriate for standard prediction models without causal interpretation
Counterfactual mode (treatment
provided):
- Estimates E[L(Y^a, g(X)) | S=0] — performance on counterfactual outcomes
- Uses both selection model P(S=0|X) and propensity model P(A=a|X, S=1)
- Appropriate when the prediction target is a counterfactual outcome
Factual Transportability for AUC and Other Metrics
All tr_* functions support factual mode:
# Factual AUC
auc_factual <- tr_auc(
predictions = transport_sim$risk_score,
outcomes = transport_sim$event,
source = transport_sim$source,
covariates = transport_sim[, c("age", "biomarker", "smoking")],
analysis = "transport",
estimator = "dr",
se_method = "none"
)
print(auc_factual)
#>
#> Factual Transportable AUC Estimation
#> ---------------------------------------------
#> Analysis: transport
#> Estimator: dr
#> N target: 1224 | N source: 1276
#>
#> Estimate: 0.6009
#>
#> Naive estimate: 0.6126
# Factual sensitivity at threshold 0.3
sens_factual <- tr_sensitivity(
predictions = transport_sim$risk_score,
outcomes = transport_sim$event,
source = transport_sim$source,
covariates = transport_sim[, c("age", "biomarker", "smoking")],
threshold = 0.3,
analysis = "transport",
estimator = "dr",
se_method = "none"
)
print(sens_factual)
#>
#> Factual Transportable Sensitivity Estimate
#> ===================================
#>
#> Estimator: DR
#> Analysis: transport
#> N (source): 1276
#> N (target): 1224
#>
#> Threshold: 0.3
#> Estimate: 0.5573
#> Naive estimate: 0.4538References for Factual Transportability
The factual transportability approach is based on:
Steingrimsson, J. A., et al. (2023). “Transporting a Prediction Model for Use in a New Target Population.” American Journal of Epidemiology, 192(2), 296-304. doi:10.1093/aje/kwac128
Li, S., et al. (2023). “Efficient estimation of the expected prediction error under covariate shift.” Biometrics, 79(1), 295-307. doi:10.1111/biom.13583
Bootstrap Standard Errors
For inference, use bootstrap standard errors:
mse_with_se <- tr_mse(
predictions = transport_sim$risk_score,
outcomes = transport_sim$event,
treatment = transport_sim$treatment,
source = transport_sim$source,
covariates = transport_sim[, c("age", "biomarker", "smoking")],
treatment_level = 0,
analysis = "transport",
estimator = "dr",
se_method = "bootstrap",
n_boot = 500,
stratified_boot = TRUE # Preserve source/target ratio
)
summary(mse_with_se)
#>
#> Summary: Counterfactual Transportable MSE Estimation
#> =======================================================
#>
#> Call:
#> tr_mse(predictions = transport_sim$risk_score, outcomes = transport_sim$event,
#> treatment = transport_sim$treatment, source = transport_sim$source,
#> covariates = transport_sim[, c("age", "biomarker", "smoking")],
#> treatment_level = 0, analysis = "transport", estimator = "dr",
#> se_method = "bootstrap", n_boot = 500, stratified_boot = TRUE)
#>
#> Settings:
#> Mode: Counterfactual
#> Analysis type: transport
#> Estimator: dr
#> Treatment level: 0
#> Target sample size: 1224
#> Source sample size: 1276
#>
#> Results:
#> Estimator Estimate SE CI_lower CI_upper
#> Transportable 0.2219 0.008787 0.2055 0.2404
#> Naive 0.2178 NA NA NA
#>
#> Difference (Transportable - Naive): 0.0041
confint(mse_with_se)
#> 2.5% 97.5%
#> tr_mse 0.205452 0.2403893The stratified_boot = TRUE option (default) ensures that
bootstrap samples preserve the ratio of source to target observations,
which is recommended for transportability analysis.
Key Assumptions
The transportability estimators rely on several assumptions:
1. Consistency in the Source and Target Populations.
For all individuals , we have if .
The observed outcome equals the potential outcome under the received treatment. Implies no interference and well-defined treatments.
2. Conditional Exchangeability in the Source Population (Trial)
Treatment is randomized in the source population, so there is no confounding between treatment and outcome given covariates.
3. Positivity of Treatment in the Source Population (Trial)
In the source population, the treatment level must have positive probability (guaranteed by randomization).
When to Use Each Function
| Scenario | Function | Analysis |
|---|---|---|
| Single population, observational data |
cf_mse(), cf_auc(),
cf_calibration()
|
- |
| Source (RCT) to target, outcomes in source only |
tr_mse(), tr_auc(),
tr_calibration()
|
"transport" |
| Source + target, outcomes in both |
tr_mse(), tr_auc(),
tr_calibration()
|
"joint" |
Comparison with cf_* Functions
The cf_* functions (counterfactual) are for
single-population analysis:
- Use observational data from one population
- Adjust for confounding between treatment and outcome
- Equivalent to “Observational (OBS)” estimators in Voter et al.
The tr_* functions (transportability) are for
two-population analysis:
- Transport from source to target population
- Leverage randomization in source (if RCT)
- Adjust for differences in covariate distributions
References
Voter, S. R., et al. (2025). “Transportability of machine learning-based counterfactual prediction models with application to CASS.” Diagnostic and Prognostic Research, 9(4). doi:10.1186/s41512-025-00201-y
Boyer, C. B., Dahabreh, I. J., & Steingrimsson, J. A. (2025). “Estimating and evaluating counterfactual prediction models.” Statistics in Medicine, 44(23-24), e70287. doi:10.1002/sim.70287
Steingrimsson, J. A., et al. (2023). “Transporting a Prediction Model for Use in a New Target Population.” American Journal of Epidemiology, 192(2), 296-304. doi:10.1093/aje/kwac128
Li, S., et al. (2023). “Efficient estimation of the expected prediction error under covariate shift.” Biometrics, 79(1), 295-307. doi:10.1111/biom.13583
Dahabreh, I. J., Robertson, S. E., Tchetgen, E. J., Stuart, E. A., & Hernán, M. A. (2019). “Generalizing causal inferences from randomized trials: counterfactual and graphical identification.” Biometrics, 75(2), 685-694.